Abiotic Factors
NO2
Year Average
nitrogen <- metadata[, lapply(.SD, mean, na.rm=TRUE),
by=c("Location", "Year"),
.SDcols=c("NO2")][order(Location)]
set(nitrogen, j = "NO2", value = round(nitrogen$NO2, 3))Graph
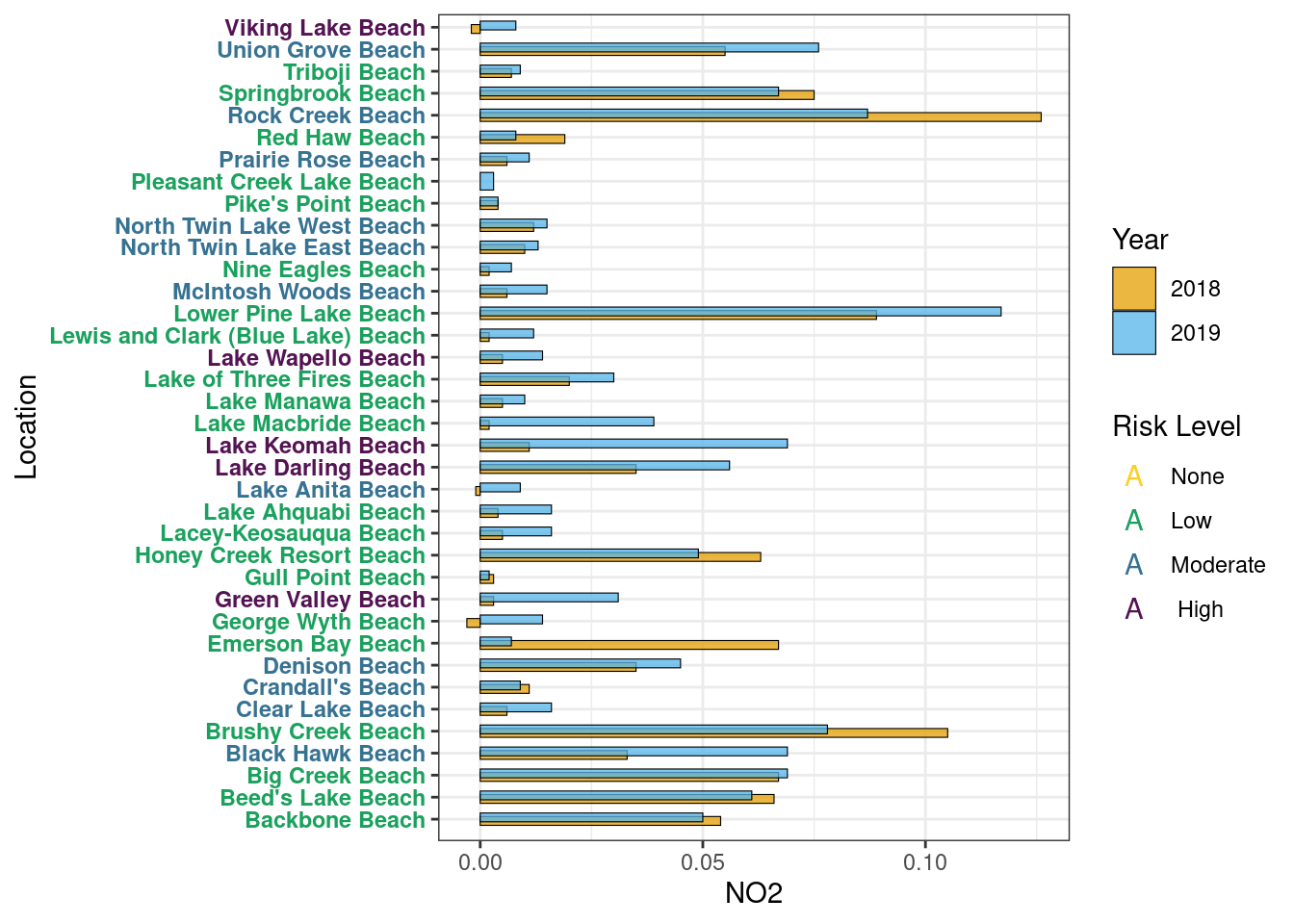
Table
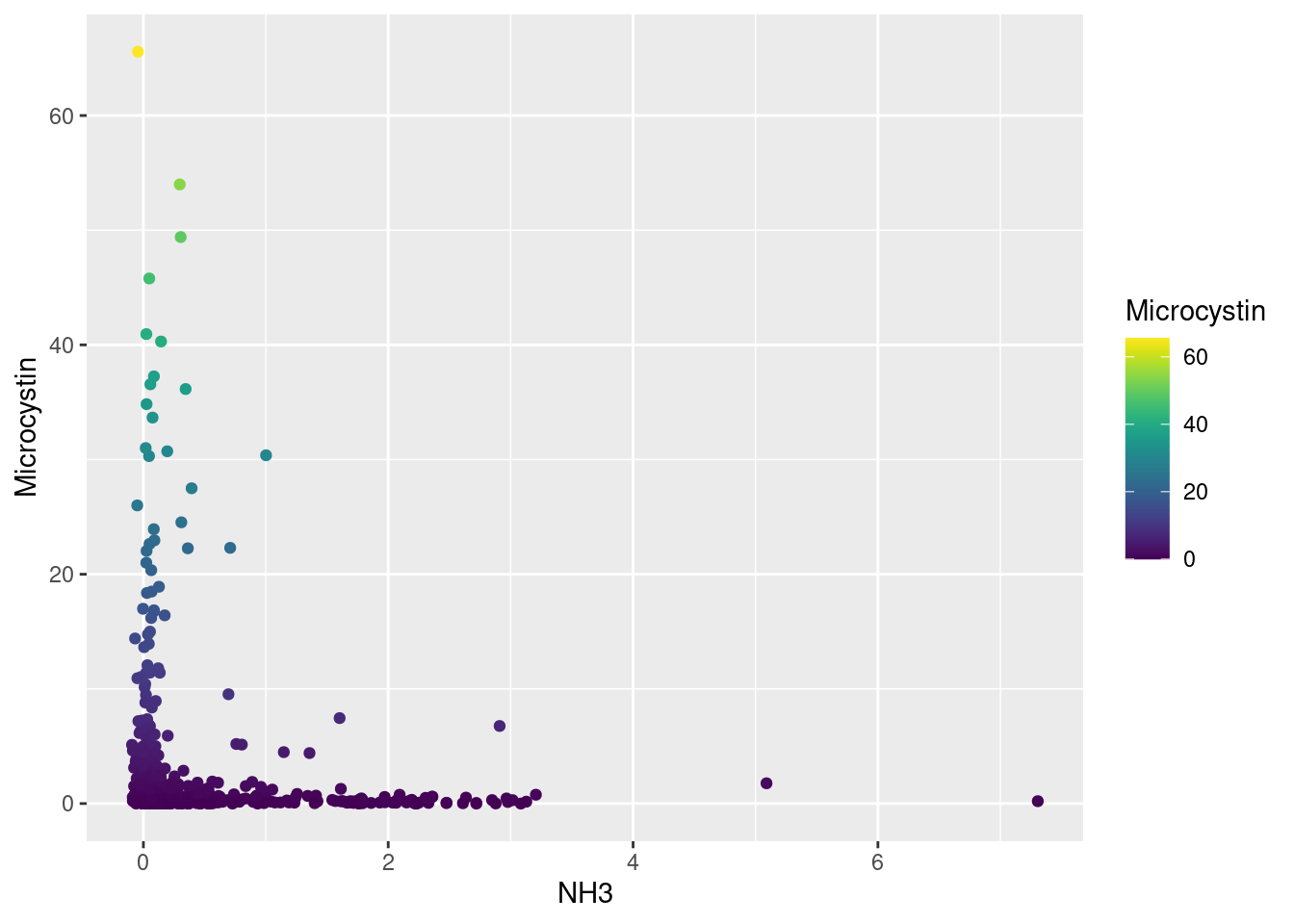
Microcystin Correlation

NH3
Year Average
NH3 <- metadata[, lapply(.SD, mean, na.rm=TRUE),
by=c("Location", "Year"),
.SDcols=c("NH3")][order(Location)]
set(NH3, j = "NH3", value = round(NH3$NH3, 3))Graph
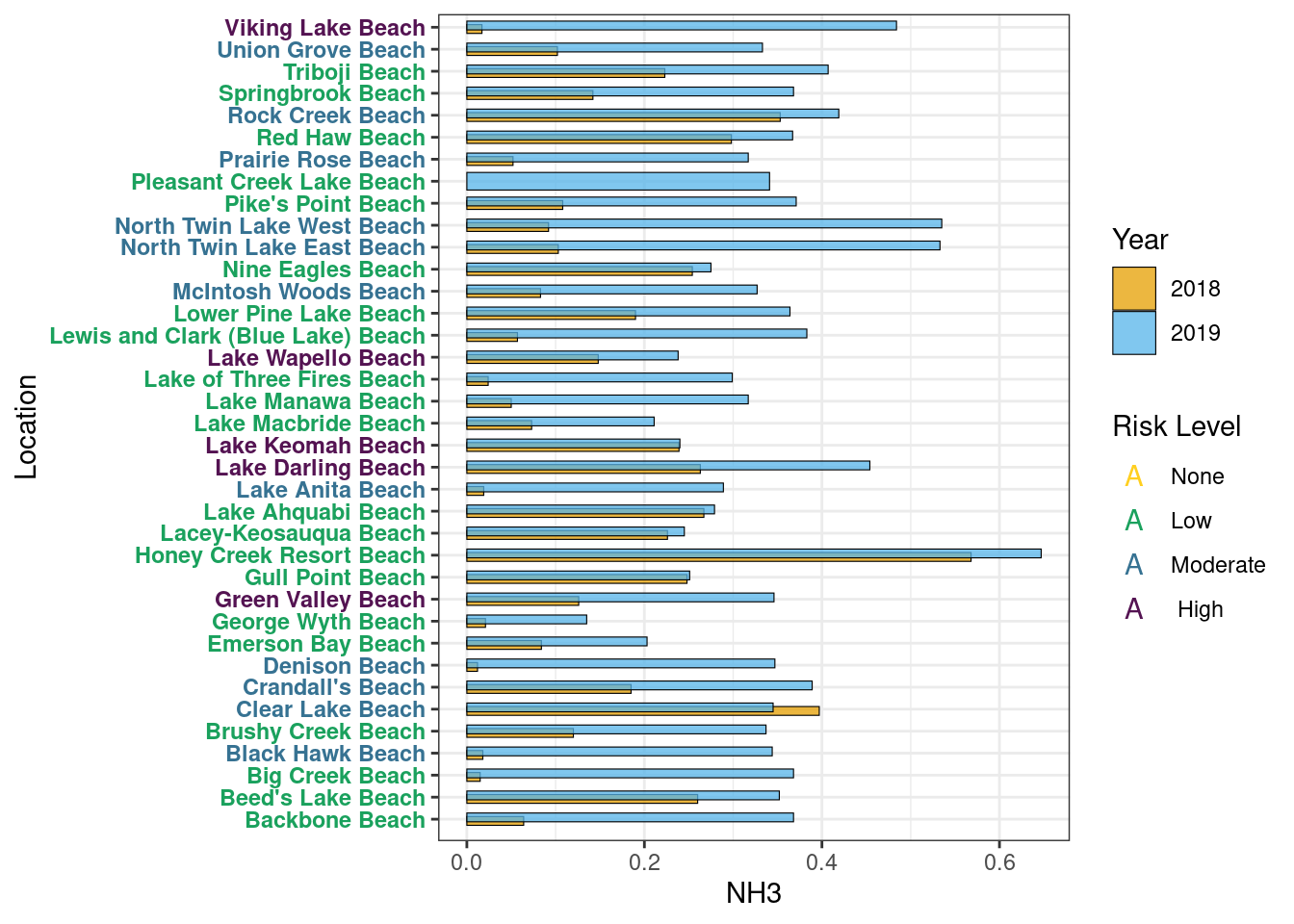
Table
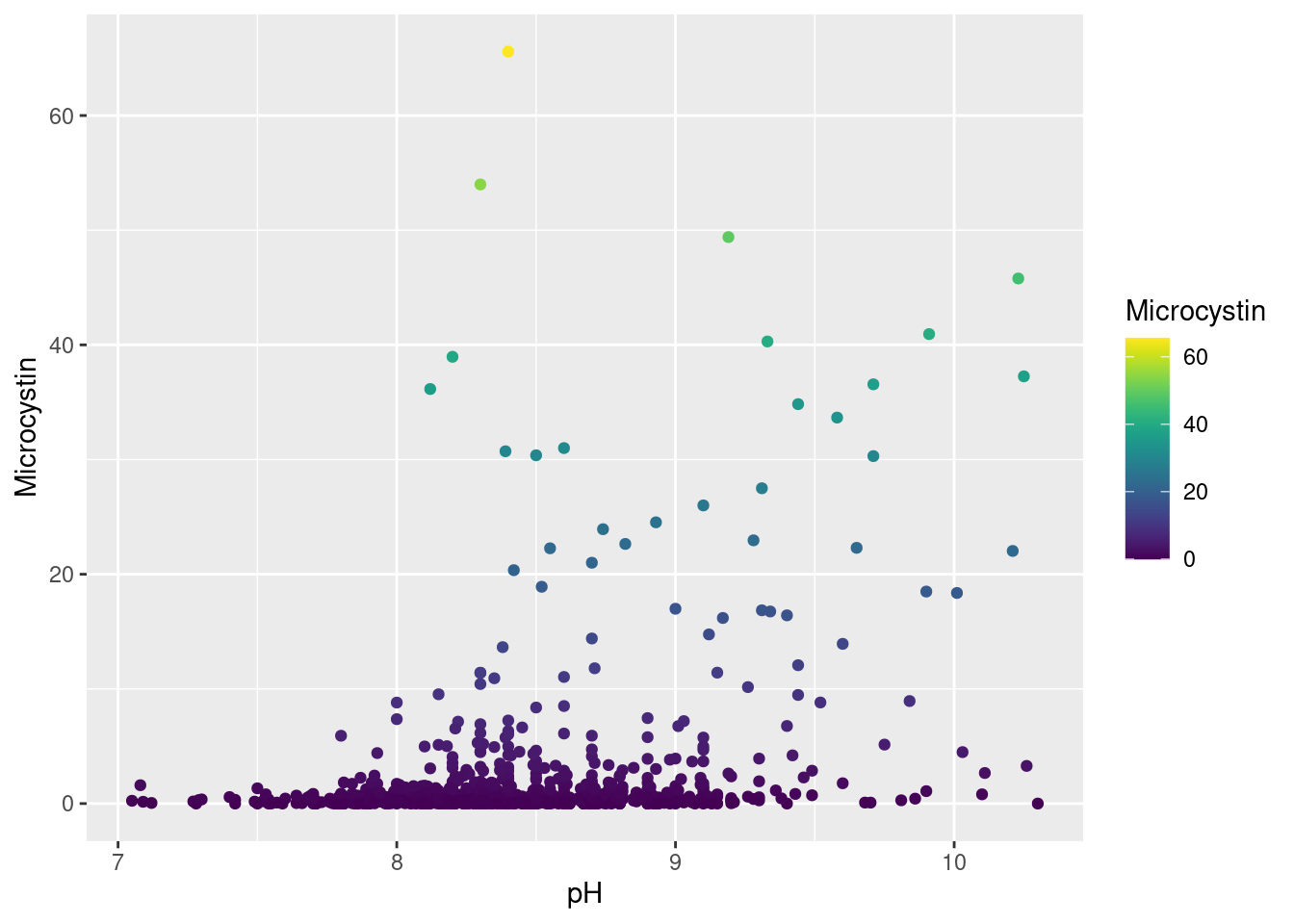
Microcystin Correlation
pH
Year Average
pH <- metadata[, lapply(.SD, mean, na.rm=TRUE),
by=c("Location", "Year"),
.SDcols=c("pH")][order(Location)]
set(pH, j = "pH", value = round(pH$pH, 2))Graph

Table
Microcystin Correlation
Cl
Year Average
Cl <- metadata[, lapply(.SD, mean, na.rm=TRUE),
by=c("Location", "Year"),
.SDcols=c("Cl")][order(Location)]
set(Cl, j = "Cl", value = round(Cl$Cl, 2))Graph
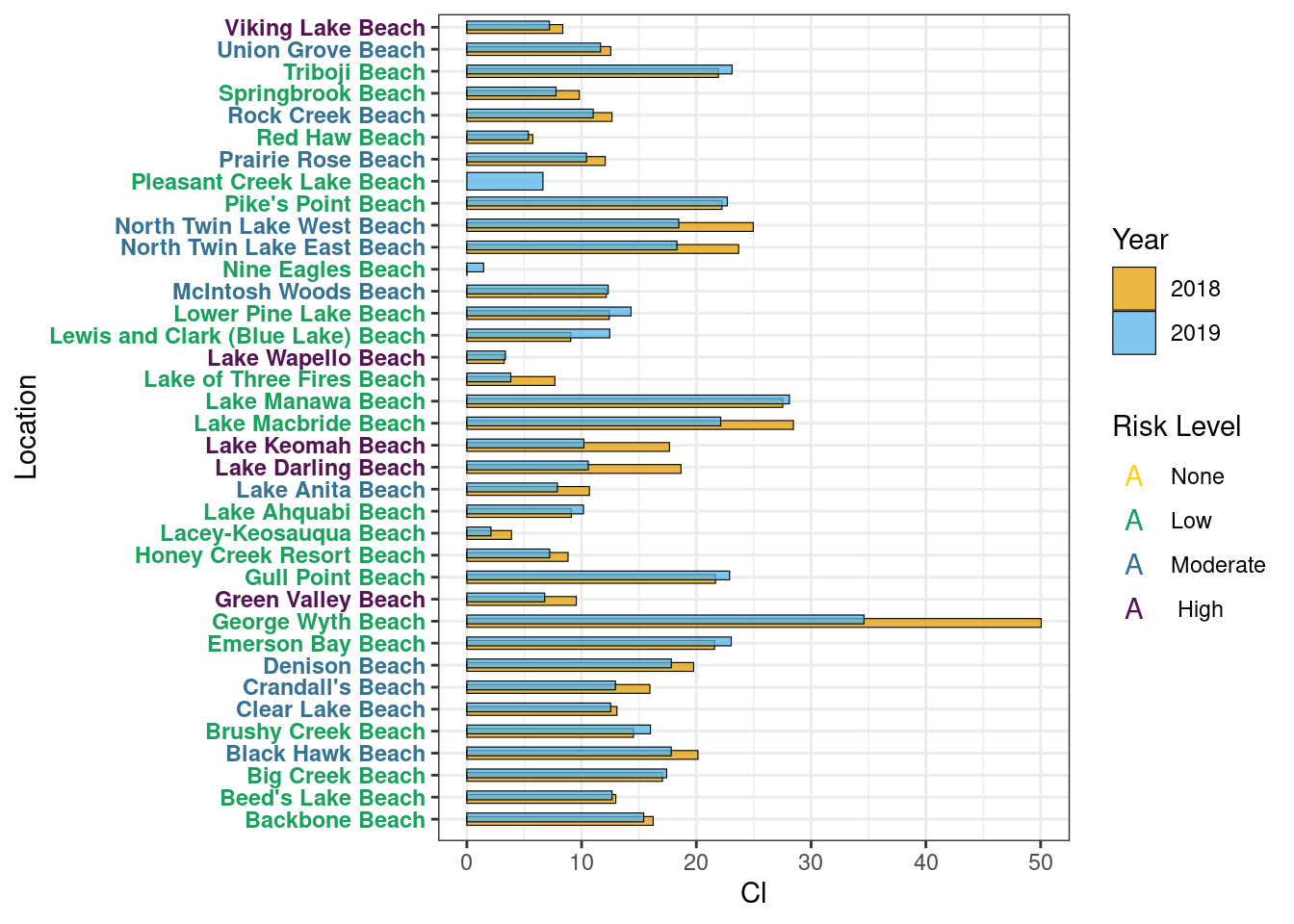
Table
Microcystin Correlation

High- & Low-Risk Lakes
Weeks of Significantly Different Variables
Microcystin Levels
2018
ggplot(sam[Year == 2018], aes(x = Week, y = Microcystin)) +
geom_bar(stat = "identity", position = "stack",
size = 0.12, width = 0.95, color = "black") +
facet_nested_wrap(.~ Lake_Risk + Location , scales = "free") +
theme_bw() +
theme(axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 10),
axis.title.x = element_text(size = 12, face = "bold"),
axis.title.y = element_text(size = 12, face = "bold"),
axis.ticks.x = element_blank(),
legend.title = element_text(size = 10, face = "bold"),
legend.text = element_text(size = 10),
legend.spacing.x = unit(0.005, "npc"),
legend.key.size = unit(3, "mm"),
legend.position="top",
panel.background = element_rect(color = "black", size = 1.5),
panel.spacing = unit(0.01, "npc"),
strip.text.x = element_text(size = 14, face = "bold"),
strip.background = element_rect(colour = "black", size = 1.4, fill = "white"),
panel.grid.major.x = element_blank()) +
scale_y_continuous(expand = expansion(mult = c(0.0037, 0.003), add = c(0, 0)), limits = c(0,70)) +
scale_x_discrete(expand = expansion(mult = 0,
add = 0.51))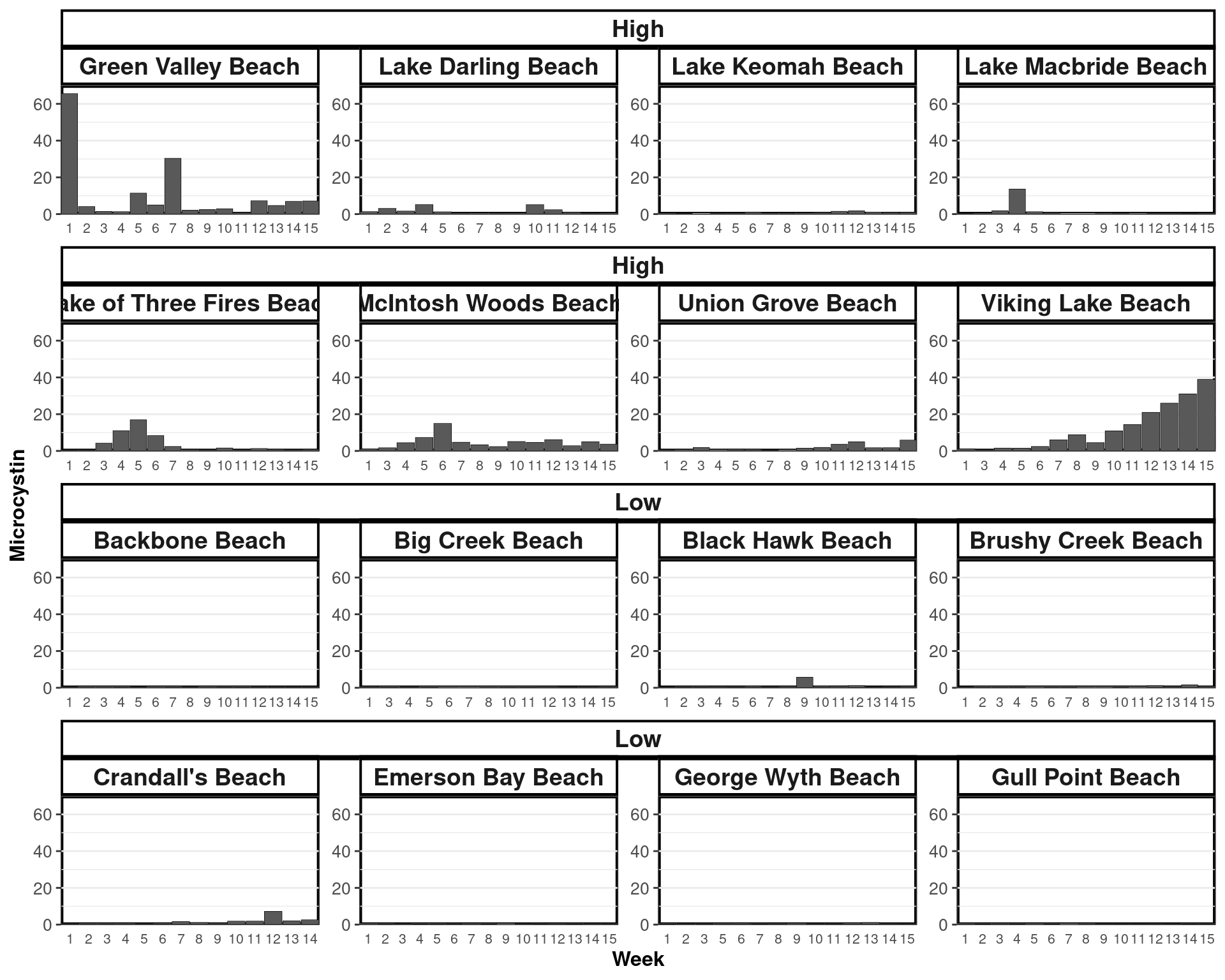
a <- data.table::copy(sam)
a[, Microcystis_x := Microcystis/max(Microcystis), by = c("Location", "Year")]
a[, Microcystin_x := Microcystin/max(Microcystin), by = c("Location", "Year")]
a <- melt.data.table(a, id.vars = c("Location", "Week", "Year", "Lake_Risk"),
measure.vars = c("Microcystin_x", "Microcystis_x"))
ggplot(a[Year == 2018], aes(x = Week, y = value,
fill = variable))+ geom_bar(stat = "identity", position = position_dodge2(padding = 3.5),
size = 0.2, color = "black", alpha = 0.85, width = 0.62) +
facet_nested_wrap(.~ Lake_Risk + Location , scales = "free") + theme_bw() +
scale_fill_manual(values = schuylR::create_palette(1 + length(unique(a$Year)))[-1],
labels = c("Microcystin", "Microcystis")) +
labs(y = "Relative Value", fill = "Variable") +
theme(axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 10),
axis.title.x = element_text(size = 12, face = "bold"),
axis.title.y = element_text(size = 12, face = "bold"),
axis.ticks.x = element_blank(),
legend.title = element_text(size = 10, face = "bold"),
legend.text = element_text(size = 10),
legend.spacing.x = unit(0.005, "npc"),
legend.key.size = unit(3, "mm"),
legend.position="top",
panel.background = element_rect(color = "black", size = 1.5),
panel.spacing = unit(0.01, "npc"),
strip.text.x = element_text(size = 14, face = "bold"),
strip.background = element_rect(colour = "black", size = 1.4, fill = "white"),
panel.grid.major.x = element_blank()) 
2019
ggplot(sam[Year == 2019], aes(x = Week, y = Microcystin)) +
geom_bar(stat = "identity", position = "stack",
size = 0.12, width = 0.95, color = "black") +
facet_nested_wrap(.~ Lake_Risk + Location , scales = "free") +
theme_bw() +
theme(axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 10),
axis.title.x = element_text(size = 12, face = "bold"),
axis.title.y = element_text(size = 12, face = "bold"),
axis.ticks.x = element_blank(),
legend.title = element_text(size = 10, face = "bold"),
legend.text = element_text(size = 10),
legend.spacing.x = unit(0.005, "npc"),
legend.key.size = unit(3, "mm"),
legend.position="top",
panel.background = element_rect(color = "black", size = 1.5),
panel.spacing = unit(0.01, "npc"),
strip.text.x = element_text(size = 14, face = "bold"),
strip.background = element_rect(colour = "black", size = 1.4, fill = "white"),
panel.grid.major.x = element_blank()) +
scale_y_continuous(expand = expansion(mult = c(0.0037, 0.003), add = c(0, 0)), limits = c(0,70)) +
scale_x_discrete(expand = expansion(mult = 0,
add = 0.51))
ggplot(a[Year == 2019], aes(x = Week, y = value,
fill = variable))+ geom_bar(stat = "identity", position = position_dodge2(padding = 3.5),
size = 0.2, color = "black", alpha = 0.85, width = 0.62) +
facet_nested_wrap(.~ Lake_Risk + Location , scales = "free") + theme_bw() +
scale_fill_manual(values = schuylR::create_palette(1 + length(unique(a$Year)))[-1],
labels = c("Microcystin", "Microcystis")) +
labs(y = "Relative Value", fill = "Variable") +
theme(axis.text.x = element_text(size = 8),
axis.text.y = element_text(size = 10),
axis.title.x = element_text(size = 12, face = "bold"),
axis.title.y = element_text(size = 12, face = "bold"),
axis.ticks.x = element_blank(),
legend.title = element_text(size = 10, face = "bold"),
legend.text = element_text(size = 10),
legend.spacing.x = unit(0.005, "npc"),
legend.key.size = unit(3, "mm"),
legend.position="top",
panel.background = element_rect(color = "black", size = 1.5),
panel.spacing = unit(0.01, "npc"),
strip.text.x = element_text(size = 14, face = "bold"),
strip.background = element_rect(colour = "black", size = 1.4, fill = "white"),
panel.grid.major.x = element_blank()) 
rm(a)
Schuyler Smith
Ph.D. Student - Bioinformatics and Computational Biology
Iowa State University. Ames, IA.